PI: David Kirchman
of: University of Delaware
dataset: Bacterial abundance Thymidine & Leucine incorporation
dates: August 10, 1992 to September 14, 1992
location: N: 12.025 S: -11.9717 W: -140.67 E: -134.95
project/cruise: EQPAC/TT011 - Fall Survey
ship: Thomas Thompson
Comments on Bacterial Data from TT007, First EqPac Survey Cruise
The data on bacterial abundance can be used to calculate
bacterial biomass (the common assumption is 20 fg C/cell) and the
incorporation rates of thymidine (TdR) and leucine (Leu) can be
used to calculate bacterial production. Any one using these
data are encouraged to contact D.Kirchman (U.Delaware) for an
update on proper factors for converting incorporation rates into
bacterial production.
The depths indicate in this data set are those original
targeted for the particular casts. That is, none of them have
been changed to reflect current guesses of where the samples were
actually taken. Also, we noted some samples that were clearly
anomalous, but again we did not delete or change these. Some of
these anomalous values may be due to problems with the CTD
bottles, but others are clearly not.
- Dave Kirchman
Measurement of Bacterial Biomass and Production (EqPac)
Hugh W. Ducklow and David L. Kirchman
Thymidine and Leucine Incorporation
Samples from the upper 200 m were collected during hydrocases with a trace-metal-free rosette (Moss Landing) and processed immediately following collection. Short-term incorporation assays followed procedures described in Ducklow et al. (1992a). Duplicate 30 ml samples were amended with methyl- H-thymidine (New England Nuclear, sp. act. >75 Ci mmol
H-thymidine (New England Nuclear, sp. act. >75 Ci mmol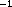 ; 10 nM final concl.) and incubated at or near in situ water temperatures in screwtop polycarbonate centrifuge tubes in chilled water bath incubators. Following incubation periods of ca. 1--3 h, the incubation was terminated with the addition of 0.5 % formalin. To measure nonspecific incorporation, these samples were filtered onto Sartorius cellulose nitrate membranes (0.22 µm pore size, extracted by rinsing the filters over a vacuum three times with ice-cold 5 % trichloroacetic acid (TCA) and three times with 80 % ethanol, as suggested in Wicks and Robarts (1988). To measure incorporation into DNA only, separate parallel samples were extracted in 0.25 n NaOH (final conc.) and chilled on ice. These samples were stored on ice for up to 48 hours, then neutralized with ice-cold 100 % TCA (final conc. 20 %), and filtered onto 22 mm dia. 0.2 µm cellulose nitrate membrane filters. Finally the samples wre extracted on the filter holders by rinsing three times each with 50 % chloroform-phenol (a 1:1 c/c mixture of liquified phenol and chloroform) and with 80 % ethanol to purify the labelled DNA (Wicks and Robarts, 1987). Zero-time controls were subtracted to correct for adsorption and other abiotic effects. The cellulose nitrate filters were packed tightly into 7 ml glass scintillation vials and dissolved in 1.0 ml of ethyl acetate, prior to addition of Ultima Gold biodegradeable scintillation cocktail (Packard). Samples were counted aboard ship on the T.G. Thompson scintillation counter.
; 10 nM final concl.) and incubated at or near in situ water temperatures in screwtop polycarbonate centrifuge tubes in chilled water bath incubators. Following incubation periods of ca. 1--3 h, the incubation was terminated with the addition of 0.5 % formalin. To measure nonspecific incorporation, these samples were filtered onto Sartorius cellulose nitrate membranes (0.22 µm pore size, extracted by rinsing the filters over a vacuum three times with ice-cold 5 % trichloroacetic acid (TCA) and three times with 80 % ethanol, as suggested in Wicks and Robarts (1988). To measure incorporation into DNA only, separate parallel samples were extracted in 0.25 n NaOH (final conc.) and chilled on ice. These samples were stored on ice for up to 48 hours, then neutralized with ice-cold 100 % TCA (final conc. 20 %), and filtered onto 22 mm dia. 0.2 µm cellulose nitrate membrane filters. Finally the samples wre extracted on the filter holders by rinsing three times each with 50 % chloroform-phenol (a 1:1 c/c mixture of liquified phenol and chloroform) and with 80 % ethanol to purify the labelled DNA (Wicks and Robarts, 1987). Zero-time controls were subtracted to correct for adsorption and other abiotic effects. The cellulose nitrate filters were packed tightly into 7 ml glass scintillation vials and dissolved in 1.0 ml of ethyl acetate, prior to addition of Ultima Gold biodegradeable scintillation cocktail (Packard). Samples were counted aboard ship on the T.G. Thompson scintillation counter.
 H-leucine incorporation was estimated in parallel incubations of samples inoculated with 0.5 nM
H-leucine incorporation was estimated in parallel incubations of samples inoculated with 0.5 nM  H-leuchine (New England Nuclear, sp. act. 153 Ci mmol
H-leuchine (New England Nuclear, sp. act. 153 Ci mmol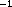 ) and 10 nM unlabeled leucine (Kirchman et al., 1985), for a final leucine concentration (hot plus cold) of 10.5 nM 30 ml leucine samples were filtered onto replicate 22 mm dia. 0.22 µm cellulose nitrate filters and extracted with ice cold 5 % TCA and ethanol as described above.
) and 10 nM unlabeled leucine (Kirchman et al., 1985), for a final leucine concentration (hot plus cold) of 10.5 nM 30 ml leucine samples were filtered onto replicate 22 mm dia. 0.22 µm cellulose nitrate filters and extracted with ice cold 5 % TCA and ethanol as described above.
Bacterial Abundance and Biomass
Samples for estimation of bacterial abundance and biovolume (20--100 ml, depending on depth) were preserved with particle-free 1.0 % glutaraldehyde then filtered within 24 h onto black Poretics polycarbonate filters (0.2 µm pore size), stained with acridine orange (Hobbie et al., 1977) and mounted in Cargille Type A immersion oil on slides and stored frozen until examination. Samples for microscopy were not replicated. All samples were enumerated using a Zeiss Axiophot microscope (final magnification 1613 x). Biovolume was estimated using the 386-based Zeiss VIDAS VIDEOPLAN Image Analysis system which acquired images from a Dage-MTI Nuvicon video camera connected to the Axiophot microscope through a Dage gen-II image intensifier. In our configuration this imaging system projects 0.2 µm spheres onto an area of approximately 17 pixels. We measure length and width (D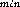 and D
and D ), perimeter and area of approximately 300 cells in each sample. The measurements are calibrated by measuring fluorescent spheres of various sizes (Polysciences Corp.). Biovolumes (V) are calculated using an algorithm (Baldwin and Bankston, 1988) which derives linear dimensions from the image analyzer's estimates of cellular perimeter, (C) and area, (A):
), perimeter and area of approximately 300 cells in each sample. The measurements are calibrated by measuring fluorescent spheres of various sizes (Polysciences Corp.). Biovolumes (V) are calculated using an algorithm (Baldwin and Bankston, 1988) which derives linear dimensions from the image analyzer's estimates of cellular perimeter, (C) and area, (A):
V = 4/3(  r
r ) + 2
) + 2 h, where (1)
_______
(C ±
h, where (1)
_______
(C ± 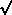 C
C -4
-4 A )
r (cell radius) = ______________ and (2)
2
A )
r (cell radius) = ______________ and (2)
2 A -
A -  r
r h (height) = ________ (3)
2r
h (height) = ________ (3)
2r
To estimate bacterial production rates from the incorporation results, conversion factors and derived empirically, loosely following the experimental design first proposed by Kirchman et al. (1982), and described more fully in Ducklow et al. (1992b). Cell volume data can be converted to biomass using various factors centering around 20 fg C 0.1 µm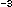 .
.
NB: These protocols closely follow methods used in JGOFS NABE by the same PI's and are quite similar to protocols in use in BATS.
Literature Cited
Baldwin, W.W. and P.W. Bankston (1988).
Measurement of live bacteria by Nomarski interference microscopy and stereologic methods as tested with macroscopic rod-shaped models. Applied and Environmental Microbiology, 54: 105--109.
Ducklow, H.W., D.L. Kirchman, H.L. Quinby, C.A. Carlson and H.G. Dam (1992a).
Response of bacterioplankton to the spring phytoplankton bloom in the eastern North Atlantic Ocean. Deep-Sea Research, (in press).
Ducklow, H.W., D.L. Kirchman and H.L. Quinby (1992b).
Bacterioplankton cell growth and macromolecular synthesis in seawater cultures during the North Atlantic spring phytoplankton bloom, May 1989. Microbial Ecology, (in press).
Hobbie, J.E., R.J. Daley and S. Jasper (1977).
Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Applied and Environmental Microbiology, 33: 1225--1228.
Kirchman, D.L., H.W. Ducklow and R. Mitchell (1982).
Estimates of bacterial growth from changes in uptake rates and biomass. Applied and Environmental Microbiology, 44: 1296--1307.
Kirchman, D.L., E. K'nees and R. Hodson (1985).
Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural waters. Applied and Environmental Microbiology, 49: 599--607.
Wicks, R.J. and R.D. Robarts (1987).
The extraction and purification of DNA labelled with [methyl- H] thymidine in aquatic bacterial production studies. Journal of Plankton Resesearch, 9: 1167--1181.
H] thymidine in aquatic bacterial production studies. Journal of Plankton Resesearch, 9: 1167--1181.
Wicks, R.J. and R.D. Robarts (1988).
Ethanol extraction requirement for purification of protein labeled with [ H] leucine in aquatic bacterial production studies. Applied and Environmental Microbiology, {f 54(12):} 3191--3193.
H] leucine in aquatic bacterial production studies. Applied and Environmental Microbiology, {f 54(12):} 3191--3193.

 ©2024 Biological and Chemical Oceanography Data Management Office.
©2024 Biological and Chemical Oceanography Data Management Office.
