Counts of seastars and chiton in quadrats and swaths, north-central California coast: Andrew Molean State Park to Manchester State Park from 2005-2014 (CHIPS project)
Project
| Contributors | Affiliation | Role |
|---|---|---|
| Gaylord, Brian | University of California - Davis: Bodega Marine Laboratory (UC Davis-BML) | Principal Investigator |
| Dawson, Michael N. | University of California-Merced (UC Merced) | Co-Principal Investigator |
| Grosberg, Richard K. | University of California-Davis (UC Davis) | Co-Principal Investigator |
| Jurgens, Laura J. | University of California - Davis: Bodega Marine Laboratory (UC Davis-BML) | Student, Contact |
| Schiebelhut, Lauren | University of California-Merced (UC Merced) | Student, Contact |
| Copley, Nancy | Woods Hole Oceanographic Institution (WHOI BCO-DMO) | BCO-DMO Data Manager |
Twenty accessible rocky intertidal sites were selected to span the ‘kill-zone’ and peripheral locations (see list of sites in Jurgens et al. submitted). Two rocky intertidal areas were sampled per site, usually one on either side of the point of entry onto the beach and separated by approximately 100 m. We used quadrats (1m2 [4 * 0.25 m^2] or 0.0625 m^2) and swaths (2 m wide transects) to estimate juvenile and adult abundance for each target species.
This data set includes counts of Pisaster ochraceus, Cryptochiton stelleri and Leptasterias sp. in quadrats and swaths along the California coast from 37.9 N to 38.9 N.
Access Restriction: Access to these data are restricted until publication of the associated manuscript, which is currently in review at PLOS ONE. [2015-02-05]
Related Datasets:
Henricia counts
Purple urchin density
Purple urchin density by depth
CHIPS02 - seastar chiton urchin counts in quadrats
Quadrats for C. stelleri, Henricia sp., Leptasterias sp., P. ochraceus:
We exhaustively searched 32-40 one-meter square quadrats per site (i.e. 16-20 per each of 2 areas), recording GPS waypoint, time, percent cover of major habitat types, and abundances and sizes of target species for each quadrat. Quadrat locations were selected by first finding one of the target habitat types - surf grass, low-zone red algae, coralline turf, cobble or boulder field, urchin pools with pits either empty or occupied, or mussel bed - selecting a starting point haphazardly, and then using a random numbers table to choose specific quadrat locations. Some sites had fewer than 32 quadrats or quadrats smaller than one-square meter due to the limited time in which we could work in suitable habitat. For mussel habitat one 0.25 m x 0.25 m quadrat was used. For Jurgens et al. (submitted) we calculated the number of each target species summed across all quadrats surveyed within each of the two areas within sites.
Swaths for C. stelleri, Henricia sp., Leptasterias sp., P. ochraceus:
Timed, GPS-tracked, 2 m wide swath transects were nested within each of the two areas within all sites. From a distance, an approximate starting point and orientation (with landmarks) for the starting transect was selected. The GPS was set to auto-record a trackpoint every 6 seconds. Transects ran from the most shoreward to the most seaward possible suitable habitat at approximately 10 m intervals along shore, particularly targeting the low zone when the tide was the lowest, with as many transects being done as permitted by the tide. The start and end of each transect was GPS waypointed and recorded. All target species were searched for across the 2 m wide swath, with intertidal zones and habitat type recorded. A waypoint was recorded for each individual target species, with any additional species found within a meter being counted and recorded for the same waypoint. When the field of view was clear some swaths were extended to 4 meters wide. For Jurgens et al. (submitted) we calculated the number of each target species summed across all swaths within each of the two areas within sites.
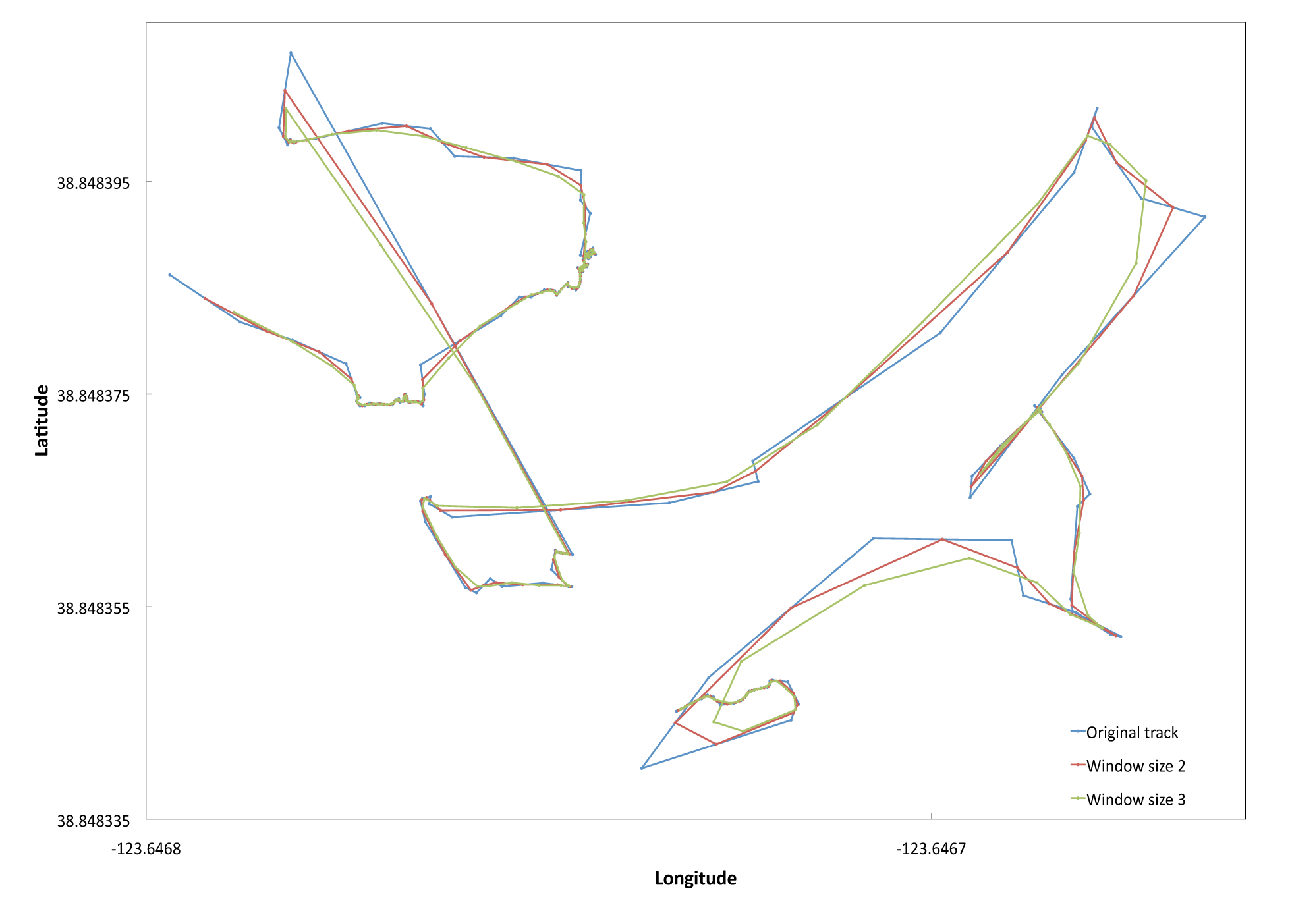
To estimate swath length we first summed the Euclidean distances between consecutive GPS trackpoints. Given the GPS had an error of +/- 3 meters we smoothed the GPS tracks by averaging across two trackpoints and then by three. We selected a window size of two with which to estimate our total transect length to adjust for the noise in our swath transects but to minimize oversmoothing. (see example figure below). We identified and removed outlying trackpoints that led to a Euclidean distance >=8 meters (likely due to temporarily poor satellite signal). The distance was then recalculated between the trackpoints immediately preceding and following the removed trackpoint. Total transect search area was calculated by multiplying the total adjusted transect length by swath width. Total transect search time was estimated using field notes and GPS waypoints and trackpoints, removing 6-second intervals where < 0.05 m were traveled (suggesting this was time during which notes were being written or tissues samples were being collected).
Swath figure: Sample GPS tracks for two transects at Iversen Point, Mendocino County. The blue line is the original unsmoothed track. The red line represents a window size of two, averaging latitudes and longitudes across each pair of consecutive trackpoints. The green line represents a window size of 3, averaging across 3 trackpoints.
BCO-DMO Processing Notes:
- combined quadrat and swath tables
- added conventional header with dataset name, PI name, version date, reference information
- added lon column
- renamed parameters to BCO-DMO standard
- sorted by latitude, high to low, then area, then date
- changed format of swath area to integer from 11 decimal places to right
- moved date column
- added area_type column
| Parameter | Description | Units |
| site | survey site name | unitless |
| lat | latitude; north is positive | decimal degrees |
| lon | longitude; east is positive | decimal degrees |
| area_type | survey area type: quadrat or swath | unitless |
| area | area of survey | m^2 |
| date | date of survey | YYYY-MM-DD |
| count_P_ochraceus | Number of Pisaster ochraceus adults found within one quadrat | unitless |
| count_C_stelleri | Number of Cryptochiton stelleri adults found within one quadrat | unitless |
| count_Leptoasterias | Number of Leptasterias adults found within an area of quadrats or swaths as defined in the dataset | unitless |
| Dataset-specific Instrument Name | GPS |
| Generic Instrument Name | Global Positioning System Receiver |
| Generic Instrument Description | The Global Positioning System (GPS) is a U.S. space-based radionavigation system that provides reliable positioning, navigation, and timing services to civilian users on a continuous worldwide basis. The U.S. Air Force develops, maintains, and operates the space and control segments of the NAVSTAR GPS transmitter system. Ships use a variety of receivers (e.g. Trimble and Ashtech) to interpret the GPS signal and determine accurate latitude and longitude. |
CalifCoast_Gaylord_CHIPS
| Website | |
| Platform | shoreside Calif_shore |
| Start Date | 2005-01-01 |
| End Date | 2014-05-15 |
| Description | Various intertidal invertebrate studies. |
Ecological & genetic recovery from a massive invertebrate die-off along the central coast of California (CHIPS)
This project is a Collaborative Research project funded by an NSF RAPID grant.
Description from NSF award abstract:
The potentially important role that larval dispersal may play in determining gene flow, distributions, and population structure of marine invertebrates remains unclear despite many hundreds of descriptive comparisons of pelagic duration and population genetic structure. This lack of clarity suggests many factors may influence population genetic structure and their interactions may be complex. Difficulties studying these factors include (under normal circumstances) distinguishing local from exogenous recruitment and therefore the true distribution of dispersal distances. For example, experiments that normally could be undertaken to explore this issue are very small scale relative to the distances that many marine taxa may disperse.
In August 2011, a large-scale natural removal experiment was initiated along a 100 km stretch of the central California coast. The PIs propose to use this rare opportunity to clarify the effects of dispersal and species interactions on marine population genetic variation and community structure. They propose to study three species that suffered very high rates of mortality: an ecosystem engineer (Strongylocentrotus purpuratus, ~100% mortality), a keystone species (Pisaster ochraceus, ~10-70% mortality), and one of its competitors (Leptasterias sp., ~100% mortality). Their objectives during this first year following the natural large-scale die-off are to:
(1) quantify the abundance and distribution of the target species at sites across the impacted range and reference sites to the south and north,
(2) develop and use genetic markers to identify the sources and dispersal distances of new recruits of P. ochraceus, Leptasterias sp., and S. purpuratus that recolonize the impacted range, and
(3) describe changes in abundance of these three species and their prey and competitors at sites throughout the impacted range.
| Funding Source | Award |
|---|---|
| NSF Division of Ocean Sciences (NSF OCE) | |
| NSF Division of Ocean Sciences (NSF OCE) |
[ table of contents | back to top ]