Proteins identified from the black smoker chimney Inferno hydrothermal vent plume meta-proteome - replicate Av1 - on the Axial seamount off the coast of Washington in 2011.
Project
| Contributors | Affiliation | Role |
|---|---|---|
| Morris, Robert | University of Washington (UW) | Principal Investigator, Contact |
| Allison, Dicky | Woods Hole Oceanographic Institution (WHOI BCO-DMO) | BCO-DMO Data Manager |
Proteins identified in the Inferno hydrothermal vent plume meta-proteome (replicate Av1). Only proteins identified by peptides with a protein probability >0.9 are listed.
These data are reported as Supplementary Table 3 and discussed in Mattes et al., 2013. (doi:10.1038/ismej.2013.113)
The FASTA information in the data was expanded to include the metadata when those FASTA headers were linked to GenBank.
Proteins that were identified in biological replicate Av2 that were not identified in biological replicate Av1. (GSO: Gamma Sulfur Oxidizer)
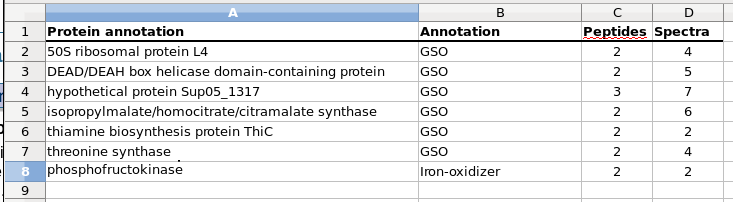
"Although fewer proteins were identified in Av2, nearly all (94%) of the proteins identified in Av2 were also identified in Av1. Differences in the total number of proteins identified in replicate samples may result from differences in the amount of biomass obtained during sample processing."
DMO notes:
Put multiple FASTA entries on separate lines
Split out one number in FASTA header for linking
Left it sorted by Total Independent Spectra column
Added linkage column
Removed commas in 'consensus annotation' column (signals database to put in new column)
Reordered columns to put KEGG last -- much longer than any other column
Seawater (~180 L) was collected from the stable hydrothermal vent plume issuing from the black smoker chimney Inferno (CTD17, 1 450 m). Whole water was transferred to clean 50 L polystyrene reservoirs and concentrated to ~230 ml with a Pellicon 2 tangential flow filtration system equipped with a 30 kDa Biomax Polyethersulfone cassette (Millipore Corporation, Billerica, MA) as described previously (Morris et al 2010). Cells were collected and concentrated in approximately 2 hours. Concentrated cells were flash frozen in liquid nitrogen and stored at -80 ºC until further processing at the University of Washington.Cell counts before and after filtration (6.9 x 1010 and 2.9 x 1010, respectively) indicate that we recovered 42% of the cells present in 180 L of hydrothermal vent plume water. Cells in the concentrated sample were divided into replicate samples (Av1 and Av2, ~115 ml each) and harvested by centrifuging at 4°C for 60 min (17,000 x g). The supernatant was discarded and cell pellets were rinsed with 100 uL of 20 mM Tris buffer pH 7.4 and stored -80°C.
Cells were lysed using a titanium sonicating micro-probe (20 sec, 10 repetitions) in a 6M urea and 50 μM ammonium bicarbonate solution. Disulfide bonds were reduced with dithiothreitol and alkylated with iodo-acetic acid. After additions of ammonium bicarbonate and methanol, 2 μg of sequence grade trypsin (Promega, Madison, WI) were added to each sample. Enzymatic digestions were incubated for 12 h at 37 oC. Resulting peptides were desalted using a macro-spin C18 column (NestGroup) following the manufacturers guidelines prior to analysis by mass spectrometry (MS).
Peptide concentrations from Axial volcano hydrothermal vent plume proteome replicates Av1 and Av2 were measured using the Thermo Scientific Nanodrop 2000/2000c, which measures the peptide bond absorbance at wavelength of 205 nm. Approximately 1 μg of peptide digest was used for each injection into the mass spectrometer. Each sample consisted of a complex mixture of peptides that were introduced into the mass spectrometer by reverse-phase chromatography using a brand new 15 cm long, 75 μm i.d. fused silica capillary column packed with C18 particles (Magic C18AQ, 100 Å, 5 μm; Michrom, Bioresources, Inc., CA) fitted with a 2 cm long, 100 μm i.d. pre-column (Magic C18AQ, 200 Å, 5μm; Michrom). Peptides were first trapped on the pre-column (5% ACN; 4 ml min-1; 7 min). Chromatographic separations were performed using an acidified (formic acid, 0.1% v/v) water-acetonitrile gradient (5-35% acetonitrile in 60 min) with a total run-time of 95 minutes.
Mass spectrometry was performed on replicates Av1 and Av2 independently using the Thermo Fisher (San Jose, Ca) linear ion trap –Orbitrap (LTQ-OT) hybrid tandem mass spectrometer. Peptides were analyzed using the data-independent Precursor Acquisition Independent from Ion Count (PAcIFIC) method (Panchaud et al 2009). Rather than requiring the mass spectrometer to select ions for fragmentation based on MS1 data, the PAcIFIC method systematically fragments ions at all m/z channels (Panchaud et al 2011). Each method file includes the full 95 minute linear HPLC gradient of 5-35% ACN over 60 minutes (see above) and covers a 21.5 m/z range using 14 contiguous, unique channels that span 2.5 m/z in the mass spectrometer. This results in a total of 45 method files per PAcIFIC analytical cycle to cover a full m/z range of 400-1400.
Protein identifications
Tandem mass spectra were interrogated against a composite database containing deduced protein sequences from lineages identified in the CTD17 clone library and lineages that are dominant in the deep ocean (background seawater). The database contained marine GSOs Candidatus Vesicomyosocius okutanii HA, Candidatus Ruthia magnifica Cm, the SUP05 metagenome (Walsh et al 2009), and SCGC AAA001-B15 (Arctic96BD-19 draft genome); the methylotrophs Methylobacter tundripaludum SV96 and Methylomicrobium alcaliphilum; iron-oxidizing bacteria Gallionella capsiferriformans ES-2 and Sideroxydans lithotrophicus ES-1; abundant lineages in seawater Candidatus Pelagibacter ubique HTCC1062; Candidatus Pelagibacter ubique HTCC1002; Ammonia-oxidizing archaea Nitrosopumilus maritimus SCM1, an uncultured marine group II (Iverson et al 2012); an incomplete hydrothermal vent metagenome (Xie et al 2011); and common contaminants. SEQUEST (v. UW2011.01.1) was used to correlate observed tandem mass spectra to peptide sequence via theoretical tandem mass spectra from the composite database described above (Eng et al 1994, Eng et al 2008). For a detailed discussion of database considerations in community proteomics see Morris et al. (2010). SEQUEST parameters included a 3.75 Da peptide mass tolerance on MS1 spectra, specifying trypsin as the enzyme, variable oxidation modification on methionine (15.9949 Da), and static modification on Cysteine residues (57.021464 Da) resulting from alkylation.
| File |
|---|
vent_proteins1.csv (Comma Separated Values (.csv), 338.90 KB) MD5:d9aa2840a9e2cfacba74d03130d9b8a9 Primary data file for dataset ID 627835 |
| Parameter | Description | Units |
| entry | data entry number; each entry number is a unit and all columns of information with the same entry number should be considered together | number |
| lat | latitude of sample collection | decimal degrees |
| lon | longitude of sample collection; West is negative | decimal degrees |
| depth | depth of sample collection | meters |
| NCBI_FASTA_link | FASTA header with embedded NCBI reference number; link is to protein page in that database; several entries with multiple links separated by commas had to be put on separate lines to enable multiple links | link |
| protein_probability | the probability that the protein identified from the peptide sequences is correct. Only proteins identified by peptides with a protein probability of >0.9 are listed | decimal number |
| num_unique_peptide | the number of unique peptide sequences identified; unique peptide is defined as a peptide -- irrespective of its length -- that exists only in one protein of a proteome of interest. The peptide sequences themselves are in the peptide sequence column | number |
| indep_spectra_tot | number of identifying peaks from the tandem mass spectrometer | number |
| peptide_seq | amino acids identified in the peptide; A=Alanine; G=Glycine; etc | text |
| consensus_annotation | describing protein X in terms of topic Y; these dominant active bacterial groups are determined by consensus annotation of identified proteins | text |
| KEGG_category | Kyoto Encyclopedia of Genes and Genomes; used to identify dominant functional classifications like metabolism or genetic information processing | text |
| Dataset-specific Instrument Name | CTD Seabird 9 plus |
| Generic Instrument Name | CTD Sea-Bird 9 |
| Dataset-specific Description | Seabird 9plus CTD with temperature and conductivity sensors. |
| Generic Instrument Description | The Sea-Bird SBE 9 is a type of CTD instrument package. The SBE 9 is the Underwater Unit and is most often combined with the SBE 11 Deck Unit (for real-time readout using conductive wire) when deployed from a research vessel. The combination of the SBE 9 and SBE 11 is called a SBE 911. The SBE 9 uses Sea-Bird's standard modular temperature and conductivity sensors (SBE 3 and SBE 4). The SBE 9 CTD can be configured with auxiliary sensors to measure other parameters including dissolved oxygen, pH, turbidity, fluorometer, altimeter, etc.). Note that in most cases, it is more accurate to specify SBE 911 than SBE 9 since it is likely a SBE 11 deck unit was used. more information from Sea-Bird Electronics |
| Dataset-specific Instrument Name | Thermo Fisher (San Jose, Ca) linear ion trap –Orbitrap (LTQ-OT) hybrid tandem mass spectrometer |
| Generic Instrument Name | Mass Spectrometer |
| Dataset-specific Description | "The hybrid Fourier Transform (FT) mass spectrometer(MS) combines a linear ion trap
MS and the Orbitrap mass analyzer. Ions generated by API
are collected in the LTQ XL followed by axial ejection to the
C-shaped storage trap which is used to store and collisionally
cool ions before injection into the orbital trap. The ions
transferred from the C-Trap are captured in the orbital trap
by rapidly increasing the electric field and the detection of
the image current from coherent ion packets takes place
after the voltages have stabilized. Signals from each of the
orbital trap outer electrodes are amplified and transformed
into a frequency spectrum by fast Fourier transformation
which is finally converted into a mass spectrum." (From Fisher Scientific)
|
| Generic Instrument Description | General term for instruments used to measure the mass-to-charge ratio of ions; generally used to find the composition of a sample by generating a mass spectrum representing the masses of sample components. |
| Dataset-specific Instrument Name | Thermo Scientific Nanodrop 2000/2000c Spectrophotometer |
| Generic Instrument Name | Spectrophotometer |
| Dataset-specific Description | Measured peptide bond absorbance at wavelength 205nm |
| Generic Instrument Description | An instrument used to measure the relative absorption of electromagnetic radiation of different wavelengths in the near infra-red, visible and ultraviolet wavebands by samples. |
TN268
| Website | |
| Platform | R/V Thomas G. Thompson |
| Start Date | 2011-08-11 |
| End Date | 2011-09-01 |
| Description | This was a two leg cruise. The National Science Foundation’s Ocean Observatory Initiative-Regional Scale Nodes cruise (August 19 – September 1, 2011) from Seattle, WA to Hydrate Ridge and Axial Seamount. The cruise began August 11 when it left the port of Seattle. |
Mixotrophic bacteria and the cryptic marine sulfur cycle: Mechanisms of carbon assimilation and sulfur oxidation in the Arctic96BD-19 GSO clade (Sulfur Oxidizers)
Description from NSF award abstract:
The ocean serves an immense reservoir of carbon, nitrogen, phosphorus, sulfur, and other elements required for all life. The active and diverse microbial populations that inhabit the oceans are responsible for mediating nutrient transformations that maintain the chemistry of seawater. A recent study identified a ubiquitous group of marine bacteria from the Arctic96BD-19 gamma-proteobacterial sulfur oxidizer (GSO) lineage that is closely related to known sulfur oxidizing species that fix inorganic carbon and oxidize sulfide in low-oxygen waters. The potential for GSOs to use reduced forms of sulfur in oxygenated waters suggests that they are a keystone species that link the marine carbon and sulfur cycles. The only known isolates from the Arctic96BD-19 lineage of GSOs are now in culture, allowing fundamental questions about their roles in carbon and sulfur cycling to be investigated. Preliminary data suggest that they use energy from the oxidation of sulfur to assimilate carbon. This project seek to address the overarching hypothesis that sulfur transformations provide the Arctic96BD- 19 lineage of GSOs with energy for organic and inorganic carbon cycling throughout the water column.
Three specific hypotheses will be tested.
1. Arctic96BD-19 cells assimilate either organic carbon or fixes inorganic carbon, depending on environmental conditions.
2. Arctic96BD-19 cells oxidize thiosulfate via formation of a tetrathionate intermediate, or using the branched thiosulfate oxidation pathway.
3. Arctic96BD-19 cells are ubiquitous sulfur oxidizers that assimilate organic and inorganic carbon through the Pacific Northwest.
A combination of laboratory growth studies of the investigator's pure cultures and comparative genomic analyses will be used. The genomic data will be used to determine whether the Arctic96BD-19 cultures possess the genetic potential to oxidize reduced sulfur to sulfate (based on possession of known core and ancillary sulfur oxidation genes), which potential oxidation pathways are used, and whether they can fix inorganic carbon. These data will help guide the physiology studies by determining the most likely forms of inorganic and organic compounds that can be utilized.
Marine bacteria are critical players in global nutrient cycles, but many of their individual and community functions in the ecosystem are not well understood. Future oceanographers will need to use cultivation-dependent and cultivation-independent methods to identify metabolic process that shape microbial communities and impact biogeochemical cycles. Student education, scientific advancement, and public awareness are all important components of this project.
| Funding Source | Award |
|---|---|
| NSF Division of Ocean Sciences (NSF OCE) |
[ table of contents | back to top ]