qPCR data from B/O Hermano Gines cruises in the CARIACO Basin Time Series Station from May to November 2014 (CariacoMetaOmics project)
Project
| Contributors | Affiliation | Role |
|---|---|---|
| Edgcomb, Virginia P. | Woods Hole Oceanographic Institution (WHOI) | Principal Investigator |
| Taylor, Gordon T. | Stony Brook University - SoMAS (SUNY-SB SoMAS) | Principal Investigator |
| Taylor, Craig | Woods Hole Oceanographic Institution (WHOI) | Co-Principal Investigator |
| Ake, Hannah | Woods Hole Oceanographic Institution (WHOI BCO-DMO) | BCO-DMO Data Manager |
Abstract
Quantitative polymerase chain reaction (qPCR) data collected from molecular samples on the CAR212 and CAR216 cruises.
All samples were collected via Niskin bottles.
DNA samples for qPCR were size fractionated into a >2.7um fraction and 0.2-2.7um fraction using a glass fiber filter pre filter in-line with a sterivex filter. DNA was preserved in lysis buffer, stored frozen, and transported back to the home laboratory. After thawing, DNA was extracted according to Frias-Lopez et al. 2008, modified by Ganesh et al. 2014. All qPCR assays were performed on an Mx3000P thermal cycler (Agilent) using the SYBRgreen filter. Assays were designed for the following genes according to the following references:
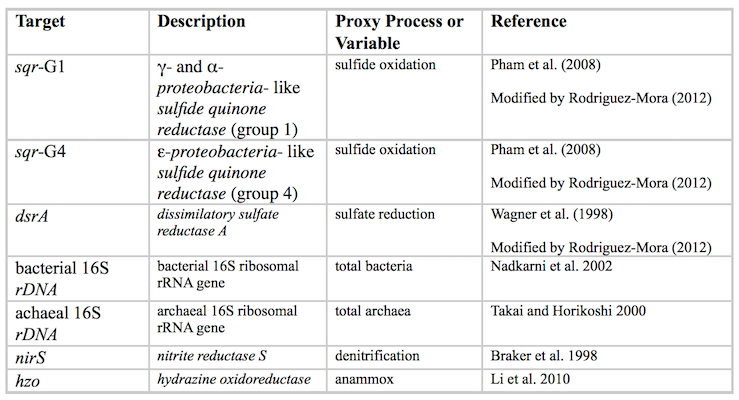
All data were normalized to volume of seawater.
DMO Notes:
-transferred each qPCR data section into separate a data object.
-reformatted the date to comply with BCO-DMO standards.
-reformatted some column names to comply with BCO-DMO standards.
| File |
|---|
qPCR.csv (Comma Separated Values (.csv), 4.10 KB) MD5:97e1ef8f4b79bed10c67a00fb932a6f4 Primary data file for dataset ID 652312 |
| Parameter | Description | Units |
| cruise_id | cariaco cruise number and leg number | unitless |
| date_start | start date; mm/dd/yyyy | unitless |
| depth | depth | meters |
| bac16S_dna | copies of the bacterial 16S gene | x10^8/L |
| bac16S_dna_sd | standard deviation of the copies of the bacterial 16S gene | x10^8/L |
| bac16S_dna_particulate | fraction of bacterial 16S genes in the greater than 2.7 um fraction | dimensionless |
| bac16S_dna_particulate_sd | standard deviation of the fraction of bacterial 16S genes in the greater than 2.7 um fraction | dimensionless |
| arch16S_dna | copies of the archaeal 16S gene | x10^8/L |
| arch16S_dna_sd | standard deviation of the copies of the archaeal 16S gene | x10^8/L |
| arch16S_dna_particulate | fraction of archaeal 16S genes in the greater than 2.7 um fraction | dimensionless |
| arch16S_dna_particulate_sd | standard deviation of the fraction of archaeal 16S genes in the greater than 2.7 um fraction | dimensionless |
| sqrG1_dna | copies of the sqrG1 gene | x10^4/L |
| sqrG1_dna_sd | standard deviation of the copies of the sqrG1 gene | x10^4/L |
| sqrG1_dna_particulate | fraction of sqrG1 genes in the greater than 2.7 um fraction | dimensionless |
| sqrG1_dna_particulate_sd | standard deviation of the fraction of sqrG1 genes in the greater than 2.7 um fraction | dimensionless |
| sqrG4_dna | copies of the sqrG4 gene | x10^4/L |
| sqrG4_dna_sd | standard deviation of the copies of the sqrG4 gene | x10^4/L |
| sqrG4_dna_particulate | fraction of sqrG4 genes in the greater than 2.7 um fraction | dimensionless |
| sqrG4_dna_particulate_sd | standard deviation of the fraction of sqrG4 genes in the greater than 2.7 um fraction | dimensionless |
| nirS_dna | copies of the nirS gene | x10^5/L |
| nirS_dna_sd | standard deviation of the copies of the nirS gene | x10^5/L |
| nirS_dna_particulate | fraction of nirS genes in the greater than 2.7 um fraction | dimensionless |
| nirS_dna_particulate_sd | standard deviation of the fraction of nirS genes in the greater than 2.7 um fraction | dimensionless |
| hzo_dna | copies of the hzo gene | x10^5/L |
| hzo_dna_sd | standard deviation of the copies of the hzo gene | x10^5/L |
| hzo_dna_particulate | fraction of hzo genes in the greater than 2.7 um fraction | dimensionless |
| hzo_dna_particulate_sd | standard deviation of the fraction of hzo genes in the greater than 2.7 um fraction | dimensionless |
| dsrA_dna | copies of the dsrA gene | x10^7/L |
| dsrA_dna_sd | standard deviation of the copies of the dsrA gene | x10^7/L |
| dsrA_dna_particulate | fraction of dsrA genes in the greater than 2.7 um fraction | dimensionless |
| dsrA_dna_particulate_sd | standard deviation of the fraction of dsrA genes in the greater than 2.7 um fraction | dimensionless |
| Dataset-specific Instrument Name | Niskin bottle |
| Generic Instrument Name | Niskin bottle |
| Dataset-specific Description | All samples were collected via Niskin bottles. |
| Generic Instrument Description | A Niskin bottle (a next generation water sampler based on the Nansen bottle) is a cylindrical, non-metallic water collection device with stoppers at both ends. The bottles can be attached individually on a hydrowire or deployed in 12, 24, or 36 bottle Rosette systems mounted on a frame and combined with a CTD. Niskin bottles are used to collect discrete water samples for a range of measurements including pigments, nutrients, plankton, etc. |
| Dataset-specific Instrument Name | Mx3000P thermal cycler (Agilent) |
| Generic Instrument Name | Thermal Cycler |
| Dataset-specific Description | All qPCR assays were performed on this thermal cycler using the SYBRgreen filter. |
| Generic Instrument Description | A thermal cycler or "thermocycler" is a general term for a type of laboratory apparatus, commonly used for performing polymerase chain reaction (PCR), that is capable of repeatedly altering and maintaining specific temperatures for defined periods of time. The device has a thermal block with holes where tubes with the PCR reaction mixtures can be inserted. The cycler then raises and lowers the temperature of the block in discrete, pre-programmed steps. They can also be used to facilitate other temperature-sensitive reactions, including restriction enzyme digestion or rapid diagnostics.
(adapted from http://serc.carleton.edu/microbelife/research_methods/genomics/pcr.html) |
CAR212_2
| Website | |
| Platform | B/O Hermano Gines |
| Start Date | 2014-05-07 |
| End Date | 2014-05-09 |
| Description | These deployments are part of the MetaOmics studies in the Cariaco Basin |
CAR216_2
| Website | |
| Platform | B/O Hermano Gines |
| Start Date | 2014-11-05 |
| End Date | 2014-11-07 |
| Description | These deployments are part of the MetaOmics studies in the Cariaco Basin. |
Genetic and Metabolic Signatures of Marine Microorganisms in Oxygen Depleted and Varying Geochemical Seascapes (CariacoMetaOmics)
Oxygen depleted water columns (ODWCs) appear to be expanding in response to global climate change. This alters trophic structure, compresses habitat and modifies geochemical cycles of major elements. Oxygen depletion can vary in intensity and duration from seasonal hypoxia to permanent anoxia. The focus of this study is a classic example of the anoxic end-member, the Cariaco Basin. The overall goal is to examine how microbial functional potential (metagenomic), activity (metatranscriptomic), taxonomic diversity (based on SSU rRNA) and the ecological/geochemical consequences (in terms of measured rates of key processes) relate along vertical oxygen/geochemical gradients and between seasons in the Cariaco Basin. This will reveal relationships between expression of particular sets of genes, environmental differences in nutrients, energy substrates and oxidant availabilities.
The objectives are to: (1) Integrate hydrographic, geochemical and microbial ecological data with metagenomic and metatranscriptomic profiles to understand regulatory and metabolic networks defining microbial community responses to environmental forcing during high and low productivity periods. This will help to understand the importance of processes, such as anaerobic oxidation of methane, utilization of redox-sensitive metals, the cryptic sulfur cycle in this ODWC, and the impacts of oxygen depletion on nitrogen transformations. (2) Determine the importance of associations between microbial eukaryotes (mEuks) and prokaryotes in this ODWC. (3) Identify "indicator" genes of known or unknown function that may be relevant to major elemental and trace gas cycling as targets for further biochemical characterization and molecular probe development, and quantify a key subset of these genes and transcripts across redox gradients using qPCR. (4) Provide a basis for developing monitoring tools using expressed genes indicative of important elemental transformations and fluxes for diagnosing the health status of natural and human engineered ecosystems. (5) Compare results with recent and ongoing studies of other ODWCs to discern shared and unique attributes of these systems.
Intellectual Merit: Previous studies of ODWCs have underscored the need for more data on microbial community structure and functionality in ODWCs, particularly biochemical rate measurements and other data on community responses to changing conditions. Better predictive models of responses of marine microbial communities and biogeochemical processes to global climate change are essential for informing future policy and management decisions. Data from an anoxic end-member ODWC like Cariaco Basin are critically needed to compare with data from other recent and ongoing studies of seasonally-depleted coastal systems and permanently-depleted deep basin and western boundary oxygen minimum zones (OMZs) to construct more skillful models. This study will advance the understanding of impacts of expanding ODWCs around the world, moving beyond assessments based only on taxonomic diversity, to yield new insights into the ecology and physiology of major microbial groups in these environments and interactions among Bacteria, Archaea and microbial eukaryotes.
Broader Impacts: The PIs and their collaborators will train one Research Associate, one postdoctoral investigator, a graduate student, and numerous undergraduates from SBU. All personnel will be trained in various aspects of microbial ecology and oceanography, with an emphasis on both traditional (e.g., microscopy) and "cutting edge" (e.g. metagenomics/transcriptomics) techniques. The PIs will also involve the Zephyr Education Foundation's marine science literacy and education program, located in Woods Hole, MA. The PIs will work with this organization to educate inner city K-12 students using local boat field trips organized by Zephyr, and lectures, and classroom laboratory exercises designed by the PIs. Additionally, this project will have broad implications for understanding how ODWCs affect marine ecosystems, and may influence future management strategies and models describing the cycling of C and N between the ocean and atmosphere.
| Funding Source | Award |
|---|---|
| NSF Division of Ocean Sciences (NSF OCE) | |
| NSF Division of Ocean Sciences (NSF OCE) |
[ table of contents | back to top ]