Holographic images from LISST-Holo system while deployed on the AUV Honey Badger (Wave Glider) during a deployment in the North Pacific gyre in 2015 (MAGI project)
Project
| Contributors | Affiliation | Role |
|---|---|---|
| Villareal, Tracy A. | University of Texas at Austin (UT Austin) | Principal Investigator, Contact |
| Wilson, Cara | National Oceanic and Atmospheric Administration - Southwest Fisheries Science Center (NOAA SWFSC ERD) | Co-Principal Investigator |
| York, Amber D. | Woods Hole Oceanographic Institution (WHOI BCO-DMO) | BCO-DMO Data Manager |
This dataset includes holographic images from a LISST-Holo Submersible Digital Holographic Camera during an AUV Honey Badger deployment in the North Pacific gyre in 2015. I Image metadata, cell counts (e.g. number of Hemiaulus cells in aggregate form), and all raw holographic images are in the RAW.zip (12GB) file.
Software for further processing the raw holographic images provided here is available at the Sequoia Scientific, Inc. LISST-Holo page The following montage and depth images can be created for every raw image using the LISST-Holo software.

The LISST-Holo Submersible Digital Holographic Camera from Sequoia Scientific, Inc. was mounted on in a tow fish deployed on a 10 m tether behind the sub-body of the AUV Honey Badger. Holographic images were acquired in bursts every 6 hours. During each burst, 15 images were taken in 5 seconds.
The software "Batch-Holo" was used to generate the biovolume from particles with an equivalent spherical diameter of 11.094-58.1 microns. See "Parameters" section for more details.
Hemiaulus and Rhizosolenia cells were identified by hand. Each image was viewed using the "Holo_Detail" software and any Hemiaulus or Rhizosolenia cells were counted.
The manual and specifications for the LISST-Holo system, and software used in this dataset are available on the instrument page. The software can be obtained from the Sequoia Scientific, Inc. LISST-Holo page.
The calculation of cells per liter was done using the following the following formula:
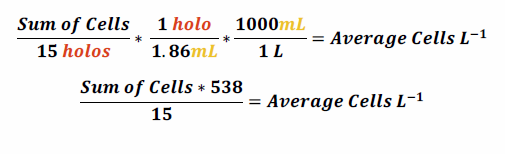
Data version 2018-01-05 replaces version 2017-10-31. The new version includes updated calculations of cells per liter. The previous version used a multiplication factor of 602 while the new version uses 538 (see Methods & Sampling section).
BCO-DMO Data Manager Processing Notes for version 2017-10-31:
* added a conventional header with dataset name, PI name, version date
* modified parameter names to conform with BCO-DMO naming conventions
* 406 values of #DIV/0! caused by zero division removed. They empty cells display as "nd" for "no data" in the served dataset.
* added CTDTime_ISO from CTD timestamp column. The CTDTime_ISO is in format yyyy-mm-ddTHH:MMZ.
* latitude,longitude,QtrLat,QtrLon rounded to 5 decimal places
* added imagename column based on the deployment number "004" and the holo image number (with 4 place 0 padding)
* added embedded with link to a thumbnail .jpg of each holographic image. Note, the thumbnails are not holographic images themselves, and are only useful for display purposes.
BCO-DMO Data Manager Processing Notes for version 2018-01-05:
* added a conventional header with dataset name, PI name, version date
* modified parameter names to conform with BCO-DMO naming conventions
* 406 values of #DIV/0! caused by zero division removed. They empty cells display as "nd" for "no data" in the served dataset.
* added CTDTime_ISO from CTD timestamp column. The CTDTime_ISO is in format yyyy-mm-ddTHH:MMZ.
* latitude,longitude,QtrLat,QtrLon rounded to 5 decimal places
* added imagename column based on the deployment number "004" and the holo image number (with 4 place 0 padding)
* added embedded with link to a thumbnail .jpg of each holographic image. Note, the thumbnails are not holographic images themselves, and are only useful for display purposes.
| File |
|---|
RAW Holographic Images filename: RAW.zip (ZIP Archive (ZIP), 11.41 GB) MD5:40ae1c9f17cac2d41f9afb5a4125b91c Raw holograms stored in .pgm format, which is a lossless compression format (see Methodology). OpenSeeIt (http://openseeit.sourceforge.net/) is a free viewer for pgm files. |
holo.csv (Comma Separated Values (.csv), 3.37 MB) MD5:b77f695d31ede318b8f224a5e5766ff9 Primary data file for dataset ID 718403 |
| Parameter | Description | Units |
| Raw_thumbnail | Reduced size image thumbnail (.jpg) of the raw holographic image for display purposes. Formatted as html with a link to larger size thumbnail. | unitless |
| Imagename_raw | Image name of the holographic raw .pgm image | unitless |
| CTDTime | Timestamp from data recorded from the CTD (UTC) in format yyyy-mm-dd HH:MM | unitless |
| CTDTime_ISO | Timestamp from data recorded in the CTD based on the ISO 8601:2004(E) standard in format YYYY-mm-ddTHH:MMZ (UTC) | unitless |
| latitude | Degrees North as recorded from the CTD | decimal degrees |
| longitude | Degrees East as recorded from the CTD | decimal degrees |
| timeStamp | Timestamp for data recorded from the LISST-Holo in UTC. Format yyyy-mm-dd HH:MM | unitless |
| Date | Date in format yyyy-mm-dd | unitless |
| holoNum | Number assigned to images taken by the LISST-Holo by the LISST-Holo | unitless |
| holoNum_difs | To track the numbering of the images. Values greater than 1 indicates a skip in numbering. Dispite the missed number there are still 15 holos/burst | unitless |
| hemiCells | Number of Hemiaulus cells identified in the holo image | individuals per image |
| hemiChains | Number of chains of Hemiaulus cells identified in the holo image | individuals per image |
| hemiAggs | Number of Hemiaulus Aggregates identified in the holo image | individuals per image |
| hemiCellsINAggs | Number of Hemiaulus cells in Aggregate form identified in the holo image | individuals per image |
| RhizoCells | Number of Rhizosolenia cells identified in the holo image | individuals per image |
| smBV | The biovolume from particles with an equvalent spherical diameter between 11.094-58.1 microns as calculated by the Holo_Batch software. "na" means the BV was due to non-phytoplankton particles (bubbles; large zooplankton; hologram errors) | microliters per liter (um/L) |
| qtrTimeStamp | Average Timestamp for the 15 holos (burst) taken 4 times a day. Format yyyy-mm-dd HH:MM | unitless |
| qtrDate | The average Date for the 15 holos (burst) taken 4 times a day. Format yyyy-mm-dd | unitless |
| qtrHCells | The sum of the Hemiaulus cells in each burst | individuals per burst |
| qtrHCellsPL | The calculated Hemiaulus cells per Liter in each burst. Calculation: (Average Hemiaulus cells in a burst) * 538 | cells per Liter |
| qtrHChains | Sum of Hemiaulus chains in each burst | individuals per burst |
| qtrHAggs | Sum of Hemiaulus Aggregates in each burst | individuals per burst |
| qtrHAggPL | Calculated number of Hemiaulus Aggregates per Liter in each burst | individuals per burst |
| qtrHAggCells | Sum of Hemiaulus cells in aggregated form in each burst. | individuals per burst |
| qtrHAggCellsPL | Calculated number of Hemiaulus cells in aggregate form per Liter in each burst | individuals per burst |
| PercHAgg | Percent of Hemiaulus cells in Aggregate form for each burst. (Total Hemiaulus cells in Aggregate form per Liter) / (Total Hemiaulus cells per Liter) | percent |
| qtrRCells | Sum of Rhizosolenia cells in each burst | individuals per burst |
| qtrRCellsPL | Calculated number of Rhizosolenia cells per Liter in each burst. Calculation: (Average Rhizonsolenia cells in a burst) * 538 | cells per Liter |
| qtrsmBV_SUM_ulL | The sum of the smBV biovolume in each burst | microliters per liter (um/L) |
| qtrsmBV_MEAN_ulL | The average smBV biovolume in each burst | microliters per liter (um/L) |
| QtrLat | Average Latitude of the Honey Badger's position for each burst | decimal degrees |
| QtrLong | Average Longitude of the Honey Badger's position for each burst | decimal degrees |
| Dataset-specific Instrument Name | GPCTD |
| Generic Instrument Name | Sea-Bird Glider Payload CTD |
| Generic Instrument Description | The GPCTD is a modular instrument for autonomous gliders. For more information see: http://www.seabird.com/glider-payload-ctd |
| Dataset-specific Instrument Name | LISST-Holo |
| Generic Instrument Name | Sequoia LISST Digital Holographic Particle Imaging System |
| Dataset-specific Description | LISST-Holo Submersible Digital Holographic Camera from Sequoia Scientific, Inc.
Specs and manual for the LISST-Holo used in this dataset:
LISST-Holo specs (PDF)
LISST-HOLO-manual-v3.0 (PDF) |
| Generic Instrument Description | Submersible Digital Holographic Particle Imaging System for measurement of large, complex flocs and biological particles. Also used for investigation of frazil ice formation and in-situ measurements of falling snow flakes. A laser beam traverses the optical path of the camera, overfilling a CCD array, creating a hologram. |
HoneyBadger2015
| Website | |
| Platform | AUV Honey Badger |
| Start Date | 2015-10-31 |
| End Date | 2015-11-04 |
| Description | The AUV Honey Badger is a Wave Glider(R) (model SV2) from Liquid Robotics. It was deployed from the Island of Hawai'i in May of 2015 as part of Mesoscale Features Aggregate Interactions (MAGI) project. The trackline is in the North Pacific Ocean and passes the ocean time-series station ALOHA.
For more information on project MAGI and a description of Honey Badger, see: http://oceanview.pfeg.noaa.gov/MAGI/ |
Long Duration AUVs as tools to explore Mesoscale feature aggregate interactions (MAGI) (MAGI)
NSF award abstract:
Remote areas of the ocean are difficult to sample for short-lived or episodic features. This project will use a new sampling platform, the Wave Glider, and provide a continuous presence in the central North Pacific gyre. The six month duration of the mission will allow repeated sampling as well as spatial coverage previously unavailable. This mission will incorporate phytoplankton specific sensors as well as a set of optical sensors that will provide information on distribution, physiology and aggregation of a unique diatom-nitrogen fixing cyanobacterium symbiosis. When completed, this program will have generated the first data sets that follow these diatom blooms over extended periods in the region. Access to this instrumentation was facilitated by the PacX challenge, an international competition to produce high quality research from long-duration autonomous vehicles in the North and South Pacific Ocean. As a result of winning that competition, the principal investigator has been awarded the use of 6 months of the Honey Badger Wave Glider time in 2014. The Wave Glider is a wave-powered surface vessel capable of extended duration missions. In order to maximize this the principal investigator will outfit the glider with advanced sensors to quantify zones of intense diatom activity and aggregation along mesoscale features in the Pacific (Project MAGI: Mesoscale feature-AGregate Interactions).
Note: This project is funded by an NSF RAPID award.
| Funding Source | Award |
|---|---|
| NSF Division of Ocean Sciences (NSF OCE) |
[ table of contents | back to top ]

